| \(Y_1=1\) | \(Y_1=2\) | \(Y_1=3\) | row total | |
|---|---|---|---|---|
| \(Y_2=1\) | 0.20 | 0.3 | 0.10 | 0.6 |
| \(Y_2=2\) | 0.15 | 0.2 | 0.05 | 0.4 |
| col. total | 0.35 | 0.5 | 0.15 | 1.0 |
Bayesian modelling
Introduction
Last compiled Tuesday Apr 15, 2025
Distribution and density function
Let \(\boldsymbol{X} \in \mathbb{R}^d\) be a random vector with distribution function \[\begin{align*} F_{\boldsymbol{X}}(\boldsymbol{x}) = \Pr(\boldsymbol{X} \leq \boldsymbol{x}) = \Pr(X_1 \leq x_1, \ldots, X_d \leq x_d). \end{align*}\]
If the distribution of \(\boldsymbol{X}\) is absolutely continuous, \[\begin{align*} F_{\boldsymbol{X}}(\boldsymbol{x}) = \int_{-\infty}^{x_d} \cdots \int_{-\infty}^{x_1} f_{\boldsymbol{X}}(z_1, \ldots, z_d) \mathrm{d} z_1 \cdots \mathrm{d} z_d, \end{align*}\] where \(f_{\boldsymbol{X}}(\boldsymbol{x})\) is the joint density function.
Mass function
By abuse of notation, we denote the mass function in the discrete case \[0 \leq f_{\boldsymbol{X}}(\boldsymbol{x}) = \Pr(X_1 = x_1, \ldots, X_d = x_d) \leq 1.\]
The support is the set of non-zero density/probability total probability over all points in the support, \[\sum_{\boldsymbol{x} \in \mathsf{supp}(\boldsymbol{X})} f_{\boldsymbol{X}}(\boldsymbol{x}) = 1.\]
Marginal distribution
The marginal distribution of a subvector \(\boldsymbol{X}_{1:k}=(X_1, \ldots, X_k)^\top\) is \[\begin{align*} F_{\boldsymbol{X}_{1:k}}(\boldsymbol{x}_{1:k}) &= \Pr(\boldsymbol{X}_{1:k} \leq \boldsymbol{x}_{1:k}) \\&= F_{\boldsymbol{X}}(x_1, \ldots, x_k, \infty, \ldots, \infty). \end{align*}\]
Marginal density
The marginal density \(f_{\boldsymbol{X}_{1:k}}(\boldsymbol{x}_{1:k})\) of an absolutely continuous subvector \(\boldsymbol{X}_{1:k}=(X_1, \ldots, X_k)^\top\) is \[\begin{align*} \int_{-\infty}^\infty \cdots \int_{-\infty}^\infty f_{\boldsymbol{X}}(x_1, \ldots, x_k, z_{k+1}, \ldots, z_{d}) \mathrm{d} z_{k+1} \cdots \mathrm{d}z_d. \end{align*}\] through integration from the joint density.
Conditional distribution
The conditional distribution function of \(\boldsymbol{Y}\) given \(\boldsymbol{X}=\boldsymbol{x}\), is \[\begin{align*} f_{\boldsymbol{Y} \mid \boldsymbol{X}}(\boldsymbol{y}; \boldsymbol{x}) = \frac{f_{\boldsymbol{X}, \boldsymbol{Y}}(\boldsymbol{x}, \boldsymbol{y})}{f_{\boldsymbol{X}}(\boldsymbol{x})} \end{align*}\] for any value of \(\boldsymbol{x}\) in the support of \(\boldsymbol{X}\).
Conditional and marginal for contingency table
Consider a bivariate distribution for \((Y_1, Y_2)\) supported on \(\{1,2,3\} \times \{1, 2\}\) whose joint probability mass function is given in Table 1
Calculations for the marginal distribution
The marginal distribution of \(Y_1\) is obtain by looking at the total probability for each row/column, e.g., \[\Pr(Y_1=i) = \Pr(Y_1=i, Y_2=1)+ \Pr(Y_1=i, Y_2=2).\]
- \(\Pr(Y_1=1)=0.35\), \(\Pr(Y_1=2)=0.5\), \(\Pr(Y_1=3) = 0.15\).
- \(\Pr(Y_2=1)=0.6\) and \(\Pr(Y_2=2)=0.4\)
Conditional distribution
The conditional distribution \[\Pr(Y_2 = i \mid Y_1=2) = \frac{\Pr(Y_1=2, Y_2=i)}{\Pr(Y_1=2)},\] so \[\begin{align*} \Pr(Y_2 = 1 \mid Y_1=2) &= 0.3/0.5 = 0.6 \\ \Pr(Y_2=2 \mid Y_1=2) &= 0.4. \end{align*}\]
Independence
Vectors \(\boldsymbol{Y}\) and \(\boldsymbol{X}\) are independent if \[\begin{align*} F_{\boldsymbol{X}, \boldsymbol{Y}}(\boldsymbol{x}, \boldsymbol{y}) = F_{\boldsymbol{X}}(\boldsymbol{x})F_{\boldsymbol{Y}}(\boldsymbol{y}) \end{align*}\] for any value of \(\boldsymbol{x}\), \(\boldsymbol{y}\).
The joint density, if it exists, also factorizes \[\begin{align*} f_{\boldsymbol{X}, \boldsymbol{Y}}(\boldsymbol{x}, \boldsymbol{y}) = f_{\boldsymbol{X}}(\boldsymbol{x})f_{\boldsymbol{Y}}(\boldsymbol{y}). \end{align*}\]
If two subvectors \(\boldsymbol{X}\) and \(\boldsymbol{Y}\) are independent, then the conditional density \(f_{\boldsymbol{Y} \mid \boldsymbol{X}}(\boldsymbol{y}; \boldsymbol{x})\) equals the marginal \(f_{\boldsymbol{Y}}(\boldsymbol{y})\).
Expected value
If \(\boldsymbol{Y}\) has density \(f_{\boldsymbol{Y}},\) then \[\begin{align*} \mathsf{E}\{g(\boldsymbol{Y})\} = \int g(\boldsymbol{y}) f_{\boldsymbol{Y}}(\boldsymbol{y}) \mathrm{d} \boldsymbol{y} \end{align*}\] a weighted integral of \(g\) with weight \(f_{\boldsymbol{Y}}.\)
The identity function gives the expected value \(\mathsf{E}(\boldsymbol{Y}).\)
Covariance matrix
We define the covariance matrix of \(\boldsymbol{Y}\) as \[\begin{align*} \mathsf{Va}(\boldsymbol{Y}) = \mathsf{E}\left[\left\{\boldsymbol{Y} - \mathsf{E}(\boldsymbol{Y})\right\}\left\{\boldsymbol{Y} - \mathsf{E}(\boldsymbol{Y})\right\}^\top\right], \end{align*}\] which reduces in the unidimensional setting to \(\mathsf{Va}(Y) = \mathsf{E}\{Y - \mathsf{E}(Y)\}^2 = \mathsf{E}(Y^2) - \mathsf{E}(Y)^2.\)
Affine transformations
If \(\boldsymbol{Y}\) is \(d\)-dimensional and \(\mathbf{A}\) is \(p \times d\) and \(\boldsymbol{b}\) is a \(p\) vector, then
\[\begin{align*} \mathsf{E}(\boldsymbol{AY} + \boldsymbol{b}) &= \boldsymbol{A}\mathsf{E}(\boldsymbol{Y}) + \boldsymbol{b},\\ \mathsf{Va}(\boldsymbol{AY} + \boldsymbol{b}) &= \boldsymbol{A}\mathsf{Va}(\boldsymbol{Y})\boldsymbol{A}^\top. \end{align*}\]
Law of iterated expectation and variance
Let \(\boldsymbol{Z}\) and \(\boldsymbol{Y}\) be random vectors. The expected value of \(\boldsymbol{Y}\) is \[\begin{align*} \mathsf{E}_{\boldsymbol{Y}}(\boldsymbol{Y}) = \mathsf{E}_{\boldsymbol{Z}}\left\{\mathsf{E}_{\boldsymbol{Y} \mid \boldsymbol{Z}}(\boldsymbol{Y})\right\}. \end{align*}\]
The tower property gives a law of iterated variance \[\begin{align*} \mathsf{Va}_{\boldsymbol{Y}}(\boldsymbol{Y}) = \mathsf{E}_{\boldsymbol{Z}}\left\{\mathsf{Va}_{\boldsymbol{Y} \mid \boldsymbol{Z}}(\boldsymbol{Y})\right\} + \mathsf{Va}_{\boldsymbol{Z}}\left\{\mathsf{E}_{\boldsymbol{Y} \mid \boldsymbol{Z}}(\boldsymbol{Y})\right\}. \end{align*}\]
Poisson distribution
The Poisson distribution has mass \[\begin{align*} f(x)=\mathsf{Pr}(Y=x) = \frac{\exp(-\lambda)\lambda^y}{\Gamma(y+1)}, \quad x=0, 1, 2, \ldots \end{align*}\] where \(\Gamma(\cdot)\) denotes the gamma function.
The parameter \(\lambda\) of the Poisson distribution is both the expectation and the variance of the distribution, meaning \[\mathsf{E}(Y)=\mathsf{Va}(Y)=\lambda.\]
Gamma distribution
A gamma distribution with shape \(\alpha>0\) and rate \(\beta>0\), denoted \(Y \sim \mathsf{gamma}(\alpha, \beta)\), has density \[\begin{align*} f(x) = \frac{\beta^\alpha}{\Gamma(\alpha)}x^{\alpha-1}\exp(-\beta x), \qquad x \in (0, \infty), \end{align*}\] where \(\Gamma(\alpha)=\int_0^\infty t^{\alpha-1}\exp(-t)\mathrm{d} t\) is the gamma function.
Poisson with random scale
To handle overdispersion in count data, take
\[\begin{align*} Y \mid \Lambda &= \lambda \sim \mathsf{Poisson}(\lambda)\\ \Lambda &\sim \mathsf{Gamma}(k\mu, k). \end{align*}\]
The joint density of \(Y\) and \(\Lambda\) on \(\mathbb{N}=\{0,1,\ldots\} \times \mathbb{R}_{+}\) is \[\begin{align*} f(y, \lambda) &= f(y \mid \lambda)f(\lambda) \\ &= \frac{\lambda^y\exp(-\lambda)}{\Gamma(y+1)} \frac{k^{k\mu}\lambda^{k\mu-1}\exp(-k\lambda)}{\Gamma(k\mu)} \end{align*}\]
Conditional distribution
The conditional distribution of \(\Lambda \mid Y=y\) can be found by considering only terms that are function of \(\lambda\), whence \[\begin{align*} f(\lambda \mid Y=y) \stackrel{\lambda}{\propto}\lambda^{y+k\mu-1}\exp\{-(k+1)\lambda\} \end{align*}\] so \(\Lambda \mid Y=y \sim \mathsf{gamma}(k\mu + y, k+1)\).
Marginal density of Poisson mean mixture
\[\begin{align*} f(y) &= \frac{f(y, \lambda)}{f(\lambda \mid y)} = \frac{\frac{\lambda^y\exp(-\lambda)}{\Gamma(y+1)} \frac{k^{k\mu}\lambda^{k\mu-1}\exp(-k\lambda)}{\Gamma(k\mu)}}{ \frac{(k+1)^{k\mu+y}\lambda^{k\mu+y-1}\exp\{-(k+1)\lambda\}}{\Gamma(k\mu+y)}}\\ &= \frac{\Gamma(k\mu+y)}{\Gamma(k\mu)\Gamma(y+1)}k^{k\mu} (k+1)^{-k\mu-y}\\&= \frac{\Gamma(k\mu+y)}{\Gamma(k\mu)\Gamma(y+1)}\left(1-\frac{1}{k+1}\right)^{k\mu} \left(\frac{1}{k+1}\right)^y \end{align*}\] Marginally, \(Y \sim \mathsf{neg. binom}(p)\) where \(p=(k+1)^{-1}.\)
Moments of negative binomial
By the laws of iterated expectation and iterative variance, \[\begin{align*} \mathsf{E}(Y) &= \mathsf{E}_{\Lambda}\{\mathsf{E}(Y \mid \Lambda\} \\& = \mathsf{E}(\Lambda) = \mu\\ \mathsf{Va}(Y) &= \mathsf{E}_{\Lambda}\{\mathsf{Va}(Y \mid \Lambda)\} + \mathsf{Va}_{\Lambda}\{\mathsf{E}(Y \mid \Lambda)\} \\&= \mathsf{E}(\Lambda) + \mathsf{Va}(\Lambda) \\&= \mu + \mu/k. \end{align*}\] The marginal distribution of \(Y\), unconditionally, has a variance which exceeds its mean.
Change of variable formula
Consider an injective (one-to-one) differentiable function \(\boldsymbol{g}: \mathbb{R}^d \to \mathbb{R}^d,\) with inverse \(\boldsymbol{g}^{-1}.\) Then, if \(\boldsymbol{Y}=\boldsymbol{g}(\boldsymbol{X}),\) \[\begin{align*} \Pr(\boldsymbol{Y} \leq \boldsymbol{y}) = \Pr\{\boldsymbol{g}(\boldsymbol{X}) \leq \boldsymbol{y}\} = \Pr\{\boldsymbol{X} \leq \boldsymbol{x} = \boldsymbol{g}^{-1}(\boldsymbol{y})\}. \end{align*}\]
Using the chain rule, the density of \(\boldsymbol{Y}\) is \[\begin{align*} f_{\boldsymbol{Y}}(\boldsymbol{y}) = f_{\boldsymbol{X}}\left\{\boldsymbol{g}^{-1}(\boldsymbol{y})\right\} \left| \mathbf{J}_{\boldsymbol{g}^{-1}}(\boldsymbol{y})\right| = f_{\boldsymbol{X}}(\boldsymbol{x}) \left| \mathbf{J}_{\boldsymbol{g}}(\boldsymbol{x})\right|^{-1} \end{align*}\] where \(\mathbf{J}_{\boldsymbol{g}}(\boldsymbol{x})\) is the Jacobian matrix with \(i,j\)th element \(\partial [\boldsymbol{g}(\boldsymbol{x})]_i / \partial x_j.\)
Gaussian location-scale
Consider \(d\) independent standard Gaussian variates \(X_j \sim \mathsf{Gauss}(0, 1)\) for \(j=1, \ldots, d,\) with joint density function \[\begin{align*} f_{\boldsymbol{X}}(\boldsymbol{x})= (2\pi)^{-d/2} \exp \left( - \frac{\boldsymbol{x}^\top\boldsymbol{x}}{2}\right). \end{align*}\] Consider the transformation \(\boldsymbol{Y} = \mathbf{A}\boldsymbol{X}+\boldsymbol{b},\) with \(\mathbf{A}\) an invertible matrix.
Change of variable for Gaussian
- The inverse transformation is \(\boldsymbol{g}^{-1}(\boldsymbol{y}) = \mathbf{A}^{-1}(\boldsymbol{y}-\boldsymbol{b}).\)
- The Jacobian \(\mathbf{J}_{\boldsymbol{g}}(\boldsymbol{x})\) is simply \(\mathbf{A},\) so the joint density of \(\boldsymbol{Y}\) is \[\begin{align*} (2\pi)^{-d/2} |\mathbf{A}|^{-1}\exp \left\{ - \frac{(\boldsymbol{y}-\boldsymbol{b})^\top\mathbf{A}^{-\top}\mathbf{A}^{-1}(\boldsymbol{y}-\boldsymbol{b})}{2}\right\}. \end{align*}\] Since \(|\mathbf{A}^{-1}| = |\mathbf{A}|^{-1}\) and \(\mathbf{A}^{-\top}\mathbf{A}^{-1} = (\mathbf{AA}^\top)^{-1},\) we recover \(\boldsymbol{Y} \sim \mathsf{Gauss}_d(\boldsymbol{b}, \mathbf{AA}^\top).\)
Conditional distribution of Gaussian subvectors
Let \(\boldsymbol{Y} \sim \mathsf{Gauss}_d(\boldsymbol{\mu}, \boldsymbol{Q}^{-1})\) and consider the partition \[\begin{align*} \boldsymbol{Y} = \begin{pmatrix} \boldsymbol{Y}_1 \\ \boldsymbol{Y}_2\end{pmatrix}, \quad \boldsymbol{\mu} = \begin{pmatrix} \boldsymbol{\mu}_1 \\ \boldsymbol{\mu}_2\end{pmatrix}, \quad \boldsymbol{Q} = \begin{pmatrix} \boldsymbol{Q}_{11} & \boldsymbol{Q}_{12}\\ \boldsymbol{Q}_{21} & \boldsymbol{\Sigma}_{22}\end{pmatrix}, \end{align*}\] where \(\boldsymbol{Y}_1\) is a \(k \times 1\) and \(\boldsymbol{Y}_2\) is a \((d-k) \times 1\) vector for some \(1\leq k < d.\)
Then, we have the conditional distribution \[\begin{align*} \boldsymbol{Y}_1 \mid \boldsymbol{Y}_2 =\boldsymbol{y}_2 &\sim \mathsf{Gauss}_k(\boldsymbol{\mu}_1-\boldsymbol{Q}_{11}^{-1}\boldsymbol{Q}_{12}(\boldsymbol{y}_2-\boldsymbol{\mu}_2), \boldsymbol{Q}^{-1}_{11}) \end{align*}\]
Likelihood
The likelihood \(L(\boldsymbol{\theta})\) is a function of the parameter vector \(\boldsymbol{\theta}\) that gives the ‘density’ of a sample under a postulated distribution, treating the observations as fixed, \[\begin{align*} L(\boldsymbol{\theta}; \boldsymbol{y}) = f(\boldsymbol{y}; \boldsymbol{\theta}). \end{align*}\]
Likelihood for independent observations
If the joint density factorizes, \[\begin{align*} L(\boldsymbol{\theta}; \boldsymbol{y})=\prod_{i=1}^n f_i(y_i; \boldsymbol{\theta}) = f_1(y_1; \boldsymbol{\theta}) \times \cdots \times f_n(y_n; \boldsymbol{\theta}). \end{align*}\] The corresponding log likelihood function for independent and identically distributions observations is \[\begin{align*} \ell(\boldsymbol{\theta}; \boldsymbol{y}) = \sum_{i=1}^n \ln f(y_i; \boldsymbol{\theta}) \end{align*}\]
Score
Let \(\ell(\boldsymbol{\theta}),\) \(\boldsymbol{\theta} \in \boldsymbol{\Theta} \subseteq \mathbb{R}^p,\) be the log likelihood function. The gradient of the log likelihood, termed score is the \(p\)-vector \[U(\boldsymbol{\theta}) = \frac{\partial \ell(\boldsymbol{\theta})}{ \partial \boldsymbol{\theta}}.\]
Information matrix
The observed information matrix is the hessian of the negative log likelihood, \[\begin{align*} j(\boldsymbol{\theta}; \boldsymbol{y})=-\frac{\partial^2 \ell(\boldsymbol{\theta}; \boldsymbol{y})}{\partial \boldsymbol{\theta} \partial \boldsymbol{\theta}^\top}, \end{align*}\] evaluated at the maximum likelihood estimate \(\widehat{\boldsymbol{\theta}},\) so \(j(\widehat{\boldsymbol{\theta}}).\)
Expected information
Under regularity conditions, the expected information, also called Fisher information matrix, is \[\begin{align*} i(\boldsymbol{\theta}) = \mathsf{E}\left\{U(\boldsymbol{\theta}; \boldsymbol{Y}) U(\boldsymbol{\theta}; \boldsymbol{Y})^\top\right\} = \mathsf{E}\left\{j(\boldsymbol{\theta}; \boldsymbol{Y})\right\} \end{align*}\]
Note on information matrices
Information matrices are symmetric and provide information about the variability of \(\widehat{\boldsymbol{\theta}}.\)
The information of an iid sample of size \(n\)is \(n\) times that of a single observation
- information accumulates at a linear rate.
Information for the Gaussian distribution
Consider \(Y \sim \mathsf{Gauss}(\mu, \tau^{-1})\), parametrized in terms of precision \(\tau\). The likelihood contribution for an \(n\) sample is, up to proportionality, \[\begin{align*} \ell(\mu, \tau) \propto \frac{n}{2}\log(\tau) - \frac{\tau}{2}\sum_{i=1}^n(Y_i^2-2\mu Y_i+\mu^2) \end{align*}\]
Gaussian information matrices
The observed and Fisher information matrices are \[\begin{align*} j(\mu, \tau) &= \begin{pmatrix} n\tau & -\sum_{i=1}^n (Y_i-\mu)\\ -\sum_{i=1}^n (Y_i-\mu) & \frac{n}{2\tau^2} \end{pmatrix}, \\ i(\mu, \tau) &= n\begin{pmatrix} \tau & 0\\ 0 & \frac{1}{2\tau^2} \end{pmatrix} \end{align*}\] Since \(\mathsf{E}(Y_i) = \mu\), the expected value of the off-diagonal entries of the Fisher information matrix are zero.
Example: random right-censoring
Consider a survival analysis problem for independent time-to-event data subject to (noninformative) random right-censoring. We observe
- failure times \(Y_i (i=1, \ldots, n)\) drawn from \(F(\cdot; \boldsymbol{\theta})\) supported on \((0, \infty)\)
- independent binary censoring indicators \(C_i \in \{0,1\}\), with \(0\) indicating right-censoring and \(C_i=1\) observed failure time.
Likelihood contribution with censoring
If individual observation \(i\) has not experienced the event at the end of the collection period, then the likelihood contribution is \(\Pr(Y > y) = 1-F(y; \boldsymbol{\theta})\), where \(y_i\) is the maximum time observed for \(Y_i\). We write the log likelihood \[\begin{align*} \ell(\boldsymbol{\theta}) = \sum_{i: c_i=0} \log \{1- F(y_i; \boldsymbol{\theta})\} + \sum_{i: c_i=1} \log f(y_i; \boldsymbol{\theta}) \end{align*}\]
Censoring and exponential data
Suppose for simplicity that \(Y_i \sim \mathsf{expo}(\lambda)\) and let \(m=c_1 + \cdots + c_n\) denote the number of observed failure times. Then, the log likelihood and the Fisher information are \[\begin{align*} \ell(\lambda) &= \lambda \sum_{i=1}^n y_i + \log \lambda m\\ i(\lambda) &= m/\lambda^2 \end{align*}\] and the right-censored observations for the exponential model do not contribute to the information.
Example: first-order autoregressive process
Consider an \(\mathsf{AR}(1)\) model of the form \[Y_t = \mu + \phi(Y_{t-1} - \mu) + \varepsilon_t,\] where
- \(\phi\) is the lag-one correlation,
- \(\mu\) the global mean and
- \(\varepsilon_t\) is an iid innovation with mean zero and variance \(\sigma^2\).
If \(|\phi| < 1\), the process is stationary, and the variance does not increase with \(t\).
Markov property and likelihood decomposition
The Markov property states that the current realization depends on the past, \(Y_t \mid Y_1, \ldots, Y_{t-1},\) only through the most recent value \(Y_{t-1}.\) The log likelihood thus becomes \[\begin{align*} \ell(\boldsymbol{\theta}) = \ln f(y_1) + \sum_{i=2}^n f(y_i \mid y_{i-1}). \end{align*}\]
Marginal of AR(1)
The \(\mathsf{AR}(1)\) stationarity process has unconditional moments \[\mathsf{E}(Y_t) = \mu, \qquad \mathsf{Var}(Y_t)=\sigma^2/(1-\phi^2).\]
The \(\mathsf{AR}(1)\) process is first-order Markov since the conditional distribution \(f(Y_t \mid Y_{t-1}, \ldots, Y_{t-p})\) equals \(f(Y_t \mid Y_{t-1})\).
Log likelihood of AR(1)
If innovations are Gaussian, we have \[Y_t \mid Y_{t-1}=y_{t-1} \sim \mathsf{Gauss}\{\mu(1-\phi)+ \phi y_{t-1}, \sigma^2\}, \qquad t>1.\] so the log-likelihood is \[\begin{align*} &\ell(\mu, \phi,\sigma^2)= -\frac{n}{2}\log(2\pi) - n\log \sigma + \frac{1}{2}\log(1-\phi^2) \\&\quad -\frac{(1-\phi^2)(y_1- \mu)^2}{2\sigma^2} - \sum_{i=2}^n \frac{(y_t - \mu(1-\phi)- \phi y_{t-1})^2}{2\sigma^2} \end{align*}\]
Estimation of integrals
Suppose we can simulate \(B\) i.i.d. variables with the same distribution, \(x_1, \ldots, x_B\) with distribution \(F\).
We want to compute \(\mathsf{E}\{g(X)\}=\int g(x) f(x) \mathrm{d} x=\mu_g\) for some functional \(g(\cdot)\)
- \(g(x)=x\) (mean)
- \(g(x) = \mathsf{I}(x \in A)\) (probability of event)
- etc.
Vanilla Monte Carlo integration
We substitute expected value by sample average of \[\begin{align*} \widehat{\mu}_g = \frac{1}{B} \sum_{b=1}^B g(x_b). \end{align*}\]
- law of large number guarantees convergence of \(\widehat{\mu}_g \to \mu_g\) if the latter is finite.
- Under finite second moments, central limit theorem gives \[\sqrt{B}(\widehat{\mu}_g - \mu_g) \sim \mathsf{No}(0, \sigma^2_g).\]
Importance sampling
Consider density \(q\) instead with \(\mathrm{supp}(p) \subseteq \mathrm{supp}(q).\) Then, \[\begin{align*} \mathsf{E}\{g(X)\} = \int_{\mathcal{X}} g(x) \frac{p(x)}{q(x)} q(x) \mathrm{d} x \end{align*}\] and we can proceed similarly by drawing samples from \(q\).
Importance sampling estimator
An alternative Monte Carlo estimator uses the weighted average \[\begin{align*} \widetilde{\mathsf{E}}\{g(X)\} =\frac{B^{-1} \sum_{b=1}^B w_b g(x_b) }{B^{-1}\sum_{b=1}^B w_b}. \end{align*}\] with weights \(w_b = p(x_b)/q(x_b)\). The latter equal 1 on average, so one could omit the denominator without harm.
Standard errors
If the variance of \(g(X)\) is finite, we can approximate the latter by the sample variance of the simple random sample and obtain the Monte Carlo standard error of the estimator \[\begin{align*} \mathsf{se}^2[\widehat{\mathsf{E}}\{g(X)\}] = \frac{1}{B(B-1)} \sum_{b=1}^B \left[ g(x_b) - \widehat{\mathsf{E}}\{g(X)\} \right]^2. \end{align*}\]
Precision of Monte Carlo integration
We want to have an estimator as precise as possible.
- but we can’t control the variance of \(g(X)\), say \(\sigma_g^2\)
- the more simulations \(B\), the lower the variance of the mean.
- sample average for i.i.d. data has variance \(\sigma^2_g/B\)
- to reduce the standard deviation by a factor 10, we need \(100\) times more draws!
Remember: the answer is random.
Example: functionals of gamma distribution
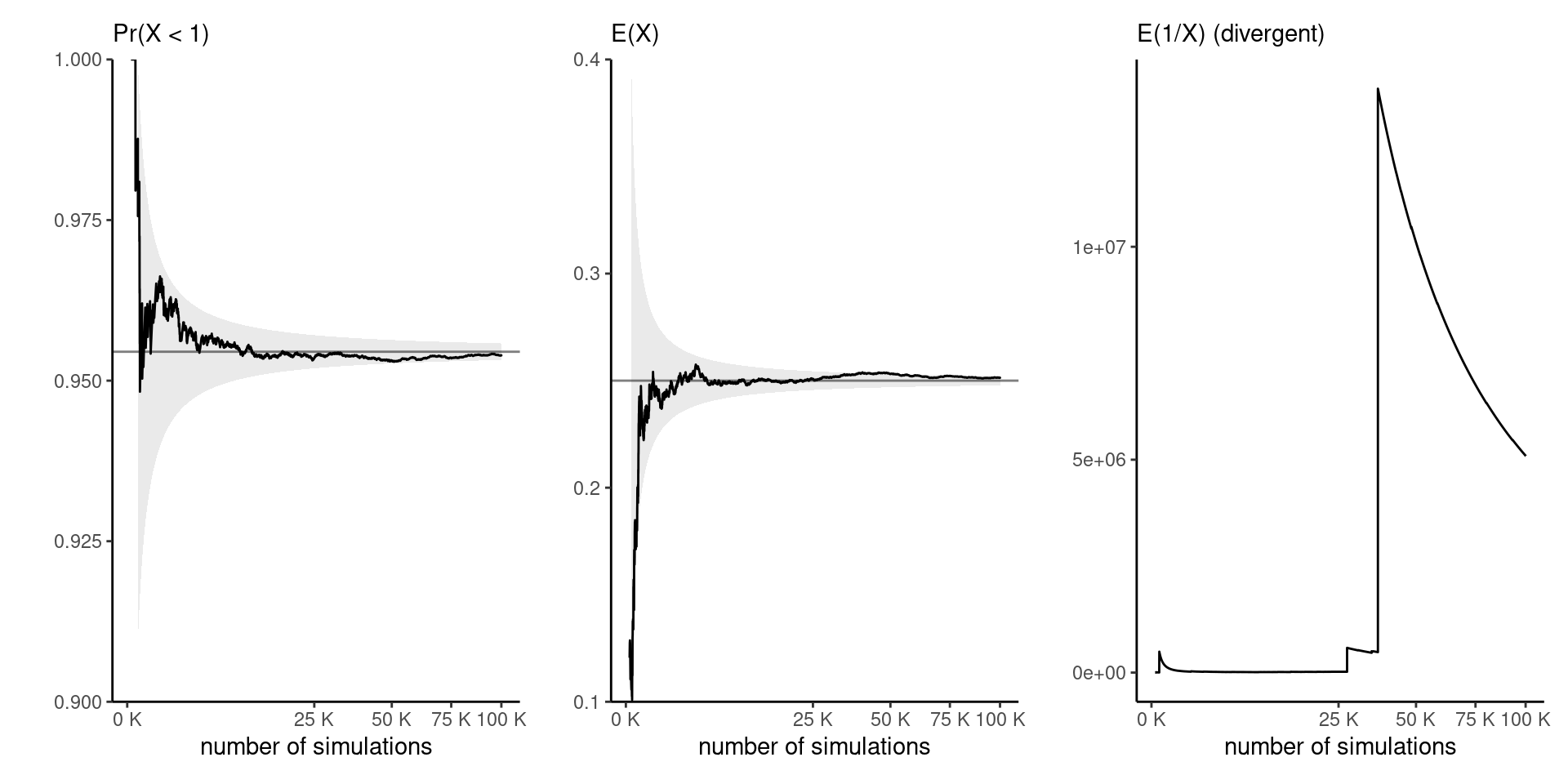
Figure 1: Running mean trace plots for \(g(x)=\mathrm{I}(x<1)\) (left), \(g(x)=x\) (middle) and \(g(x)=1/x\) (right) for a Gamma distribution with shape 0.5 and rate 2, as a function of the Monte Carlo sample size.
Recap
- We can specify distribution using hierarchies, with marginal \(\times\) conditional.
- Most density and mass functions for \(\boldsymbol{Y}\) can be identified from their support and their kernel, i.e., terms that depend on \(\boldsymbol{y}\), ignoring normalizing constants. We then match expressions.
- Expectations can be calculated analytically, or approximated via Monte Carlo simulations.
