This function computes the score test with the piecewise generalized Pareto distribution under the null hypothesis that the generalized Pareto with a single shape parameter is an adequate simplification. The score test statistic is calculated using the observed information matrix; both hessian and score vector are obtained through numerical differentiation.
Arguments
- time
excess time of the event of follow-up time, depending on the value of event
- time2
ending excess time of the interval for interval censored data only.
- event
status indicator, normally 0=alive, 1=dead. Other choices are
TRUE/FALSE(TRUEfor death). For interval censored data, the status indicator is 0=right censored, 1=event at time, 2=left censored, 3=interval censored. Although unusual, the event indicator can be omitted, in which case all subjects are assumed to have experienced an event.- thresh
a vector of thresholds
- ltrunc
lower truncation limit, default to
NULL- rtrunc
upper truncation limit, default to
NULL- type
character string specifying the type of censoring. Possible values are "
right", "left", "interval", "interval2".- weights
weights for observations
- test
string, either
"score"for the score test or"lrt"for the likelihood ratio test.- arguments
a named list specifying default arguments of the function that are common to all
elifecalls- ...
additional parameters, currently ignored
Value
a data frame with the following variables:
thresh: threshold for the generalized Pareto distributionnexc: number of exceedancesscore: score statisticdf: degrees of freedompval: the p-value obtained from the asymptotic chi-square approximation.
Details
The score test is much faster and perhaps less fragile than the likelihood ratio test: fitting the piece-wise generalized Pareto model is difficult due to the large number of parameters and multimodal likelihood surface.
The reference distribution is chi-square
Examples
# \donttest{
set.seed(1234)
n <- 100L
x <- samp_elife(n = n,
scale = 2,
shape = -0.2,
lower = low <- runif(n),
upper = upp <- runif(n, min = 3, max = 20),
type2 = "ltrt",
family = "gp")
test <- nc_test(
time = x,
ltrunc = low,
rtrunc = upp,
thresh = quantile(x, seq(0, 0.5, by = 0.1)))
print(test)
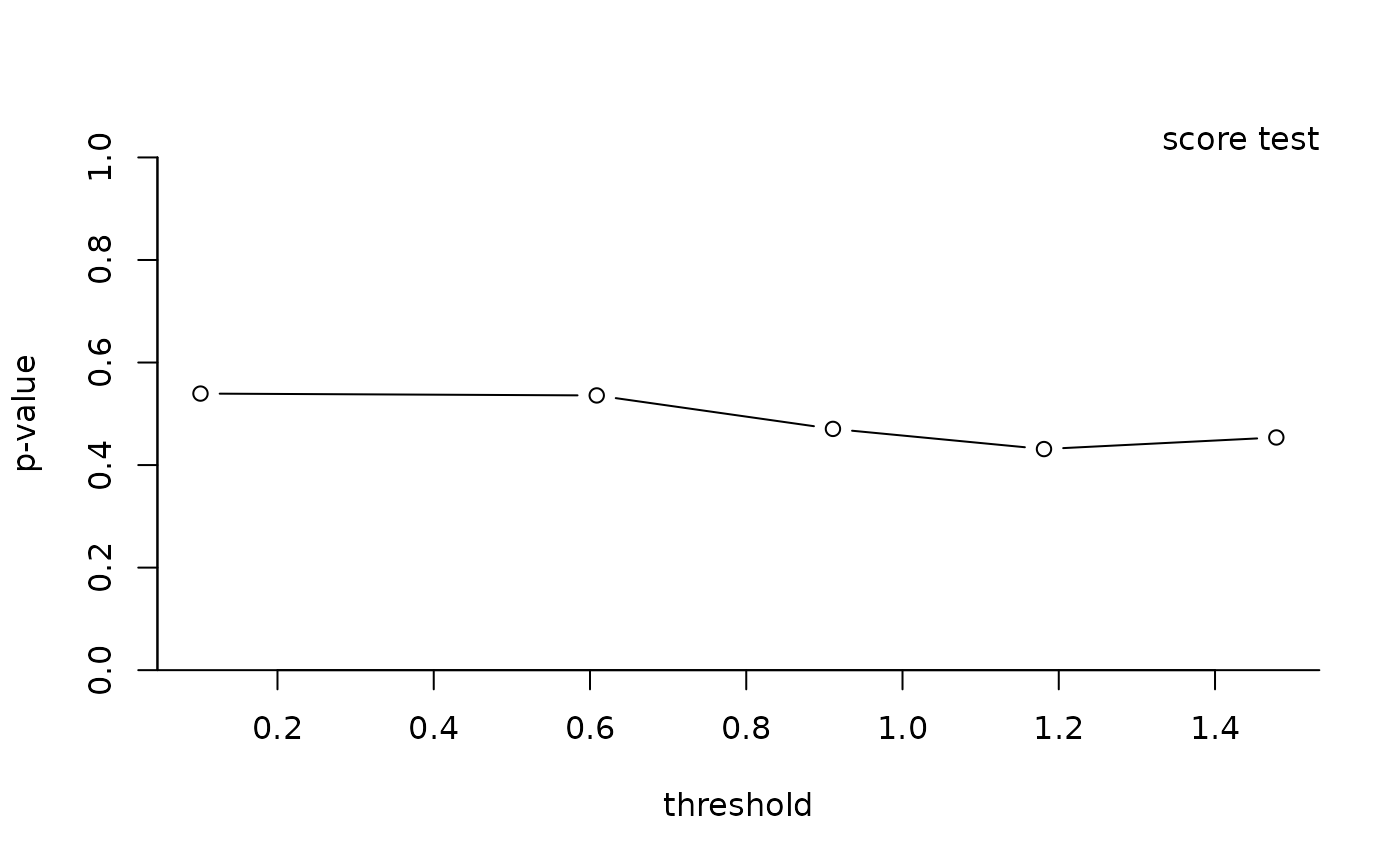
#> thresh nexc stat df pval
#> 1 0.1014320 99 4.0696252 5 0.539
#> 2 0.6086200 90 3.1330220 4 0.536
#> 3 0.9109631 80 2.5263199 3 0.471
#> 4 1.1810817 70 1.6822927 2 0.431
#> 5 1.4784588 60 0.5610536 1 0.454
plot(test)
 # }
# }