What does longevity do?
The longevity package provides a variety of numerical
routines for parametric and nonparametric models for positive data
subject to non informative censoring and truncation mechanisms. The
package includes functions to estimate various parametric model
parameters via maximum likelihood, produce diagnostic plots accounting
for survival patterns, compare nested models using analysis of deviance,
etc.
The syntax of longevity follows that of the popular
survival package, but forgoes the specification of
Surv type objects: rather, users must specify some of the
following
- the time vector
time(a left interval iftime2is provided) - the right interval
time2for interval censoring - a vector or scalar
eventindicating whether data are right, left or interval censored. The optioninterval2, for interval censoring, is useful if bothtimeandtime2vectors are provided with (potentially zero or infinite bounds) for censored observations. - the status indicator,
event, with 0 for right censored, 1 for observed event, 2 for left censored and 3 for interval censored. If omitted,eventis set to 1 for all subjects. -
ltruncandrtruncfor left and right truncation values. If omitted, they are set to 0 and \(\infty\), respectively.
The reason for specifying the ltrunc and
rtrunc vector outside of the usual arguments is to
accomodate instances where there is both interval censoring and interval
truncation; survival supports left-truncation
right-censoring for time-varying covariate models, but this isn’t really
transparent.
Example
We consider Dutch data from CBS; these data were analysed in Einmahl, Einmahl, and Haan (2019). For simplicity, we keep only Dutch people born in the Netherlands, who were at least centenarians when they died and whose death date is known.
thresh0 <- 36525
data(dutch, package = "longevity")
dutch1 <- subset(dutch, ndays > thresh0 & !is.na(ndays) & valid == "A")We can fit various parametric models accounting for the fact that
data are interval truncated. First, we create a list to avoid having to
type the name of all arguments repeatedly. These, if not provided
directly to function, are selected from the list through
arguments.
args <- with(dutch1, list(
time = ndays, # time vector
ltrunc = ltrunc, # left truncation bound
rtrunc = rtrunc, # right truncation
thresh = thresh0, # threshold (model only exceedances)
family = "gp")) # choice of parametric modelThe generalized Pareto distribution can be used for extrapolation, provided that the threshold is high enough that shape estimates are more or less stable. To check this, we can produce threshold stability plots, which display point estimates with 95% profile-based pointwise confidence intervals.
tstab_c <- tstab(
arguments = args,
family = "gp", # parametric model, here generalized Pareto
thresh = 102:108 * 365.25, # overwrites thresh
method = "wald", # type of interval, Wald or profile-likelihood
plot = FALSE) # by default, calls 'plot' routine
plot(tstab_c,
which.plot = "shape",
xlab = "threshold (age in days)")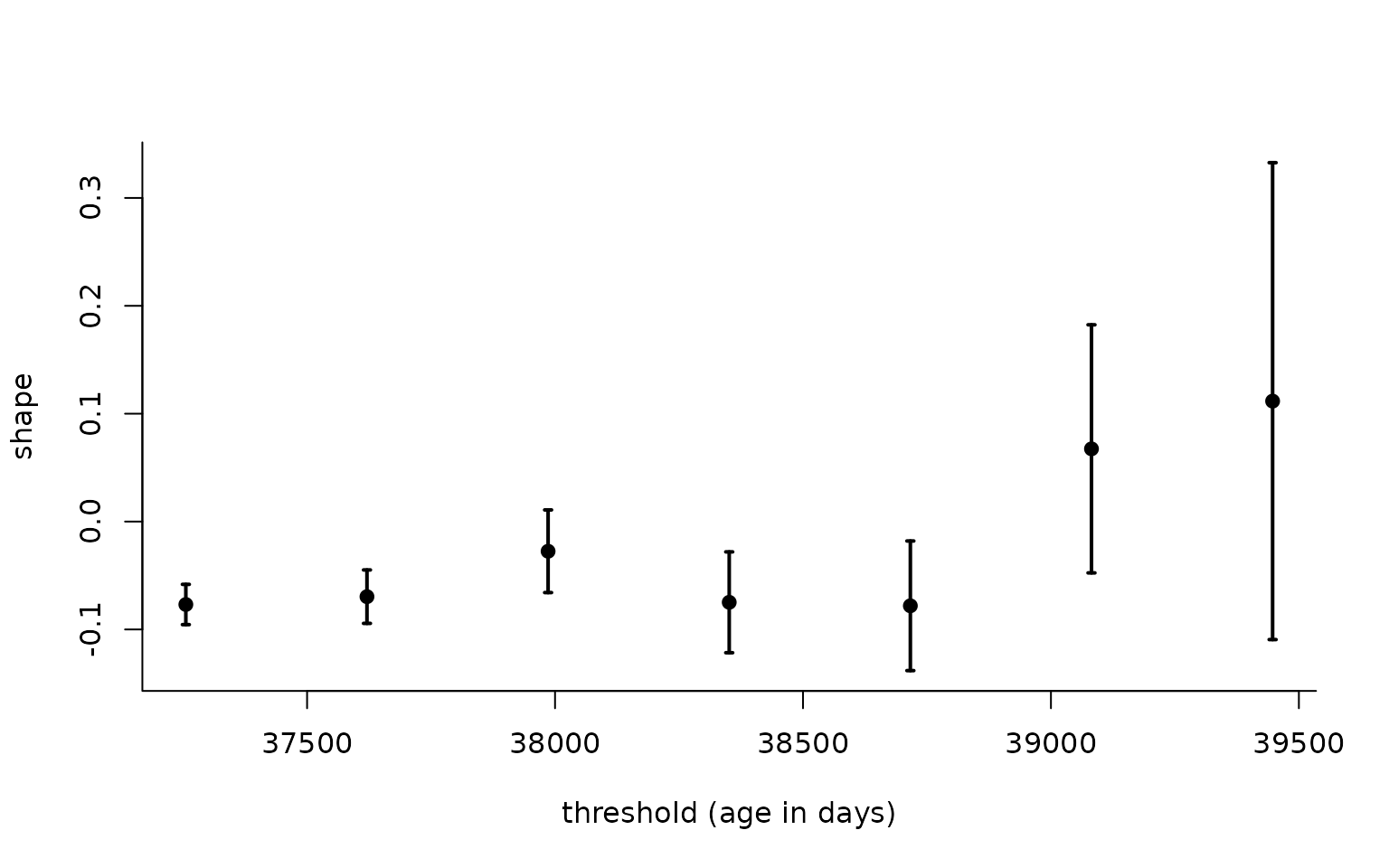
Threshold stability plot with generalized Pareto shape estimates for Dutch data as a function of threshold (in years).
We can fit various parametric models and compare them using the
anova call, provided they are nested and share the same
data. Diagnostic plots, adapted for survival data, can be used to check
goodness-of-fit. These may be computationally intensive to produce in
large samples, since they require estimation of the nonparametric
maximum likelihood estimator of the distribution function. As such, we
pick a relatively high threshold, 108 years, to reduce the computational
burden.
(m1 <- fit_elife(arguments = args,
thresh = 108 * 365.25,
family = "gp",
export = TRUE))## Model: generalized Pareto distribution.
## Sampling: interval truncated
## Log-likelihood: -631.242
##
## Threshold: 39447
## Number of exceedances: 90
##
## Estimates
## scale shape
## 412.688 0.112
##
## Standard Errors
## scale shape
## 1.414 0.113
##
## Optimization Information
## Convergence: TRUE## npar Deviance Df Chisq Pr(>Chisq)
## gp 2 1262.483 NA NA NA
## exp 1 1263.112 1 0.6287453 0.4278159
plot(m1, which.plot = "qq")
Quantile-quantile plot of generalized Pareto model to exceedances above 108 years for Dutch.