Profile likelihood for the endpoint of the generalized Pareto distribution
Source:R/endpoint.R
prof_gp_endpt.RdThis function returns the profile log likelihood over a grid of values of psi, the endpoints.
Arguments
- time
excess time of the event of follow-up time, depending on the value of event
- time2
ending excess time of the interval for interval censored data only.
- event
status indicator, normally 0=alive, 1=dead. Other choices are
TRUE/FALSE(TRUEfor death). For interval censored data, the status indicator is 0=right censored, 1=event at time, 2=left censored, 3=interval censored. Although unusual, the event indicator can be omitted, in which case all subjects are assumed to have experienced an event.- thresh
vector of thresholds
- type
character string specifying the type of censoring. Possible values are "
right", "left", "interval", "interval2".- ltrunc
lower truncation limit, default to
NULL- rtrunc
upper truncation limit, default to
NULL- weights
weights for observations
- psi
mandatory vector of endpoints at which to compute the profile
- confint
logical; if
TRUE, return alevelconfidence interval instead of a list with the profile log-likelihood components- level
numeric; the level for the confidence intervals
- arguments
a named list specifying default arguments of the function that are common to all
elifecalls- ...
additional parameters, currently ignored
Examples
set.seed(2023)
time <- relife(n = 100, scale = 3, shape = -0.3, family = "gp")
endpt <- prof_gp_endpt(
time = time,
psi = seq(max(time) + 1e-4, max(time) + 40, length.out = 51L))
print(endpt)
#> Parameter: endpoint
#> Maximum likelihood estimator: 10.77
plot(endpt)
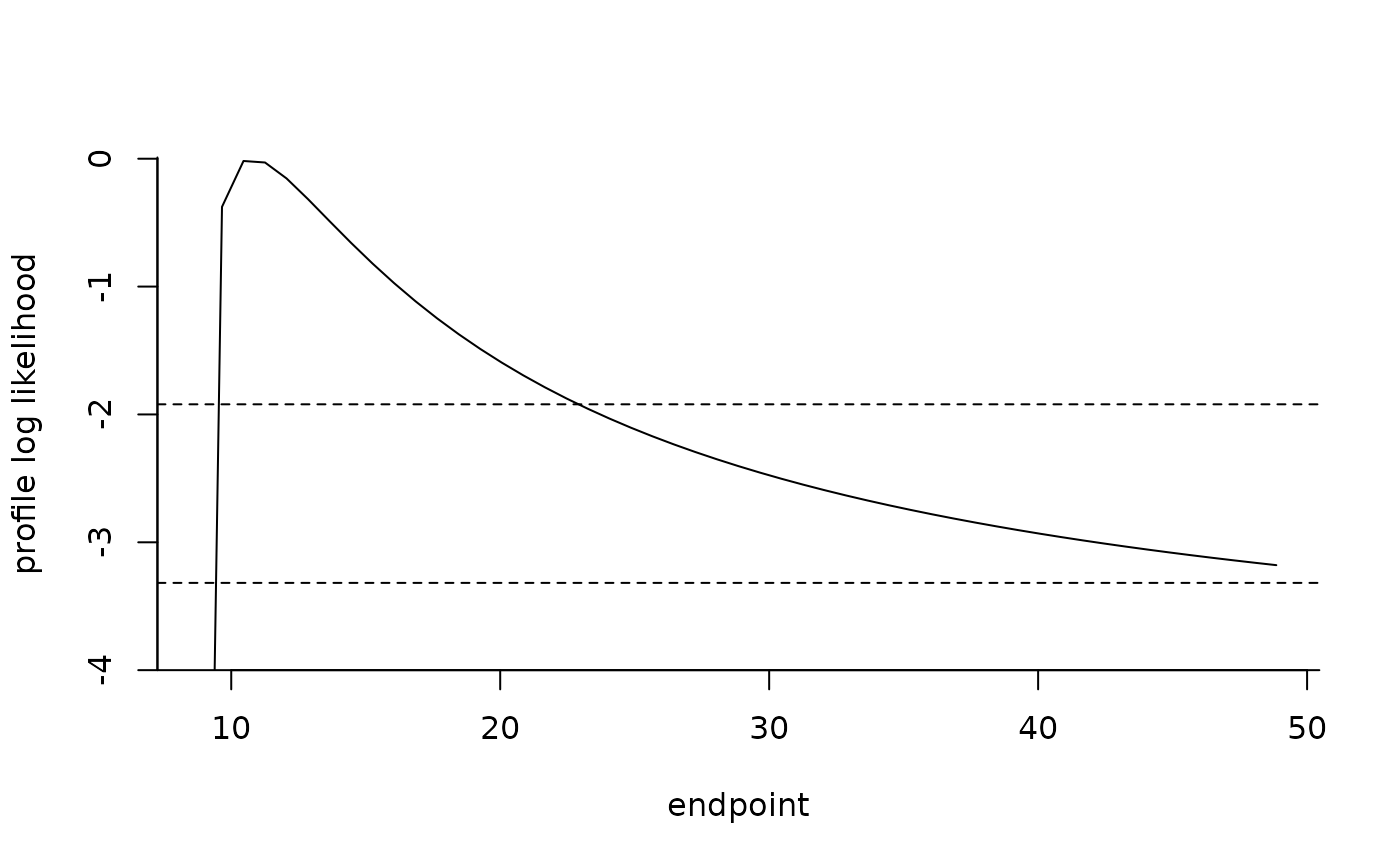 confint(endpt)
#> Estimate Lower CI Upper CI
#> 10.769592 9.168439 22.908076
confint(endpt)
#> Estimate Lower CI Upper CI
#> 10.769592 9.168439 22.908076