8.1 Bias and variance tradeoff
As we increase the penalty \(\lambda\), the values of the ridge coefficients are shrunk towards zero. The case \(\lambda \to \infty\) gives \(\hat{\boldsymbol{\beta}}_{\mathrm{ridge}}=\boldsymbol{0}_p\), whereas we retrieve the OLS estimator \(\hat{\boldsymbol{\beta}}\) when \(\lambda=0\).
The mean squared error of the ridge estimator is \[\begin{align*} \mathrm{MSE}(\hat{\boldsymbol{\beta}}_{\mathrm{ridge}}^{\lambda}) &= \sigma^2 \mathrm{tr}\left\{(\mathbf{Z}^\top\mathbf{Z} + \lambda \mathbf{I}_p)^{-1}\mathbf{Z}^\top\mathbf{Z}(\mathbf{Z}^\top\mathbf{Z} + \lambda \mathbf{I}_p)^{-1}\right\} \\&\quad + \boldsymbol{\gamma}^\top \left\{ \mathbf{Z}^\top\mathbf{Z}(\mathbf{Z}^\top\mathbf{Z} + \lambda \mathbf{I}_p)^{-1} + \mathbf{I}_p \right\} \left\{ (\mathbf{Z}^\top\mathbf{Z} + \lambda \mathbf{I}_p)^{-1}\mathbf{Z}^\top\mathbf{Z} + \mathbf{I}_p \right\}\boldsymbol{\gamma} \end{align*}\]
If we knew the true data generating mechanism (i.e. the \(\boldsymbol{\gamma}\) vector and \(\sigma^2\)), we could compute exactly the mean squared error (MSE) of the model and find the optimal bias-variance tradeoff that minimizes MSE. This is illustrated below in an artificial example. As \(\lambda \to \infty\), the bias grows and dominates MSE.
# Function to compute MSE of ridge estimator
mse_ridge <- function(gamma, lambda, Z, sigmasq = 1){
ZtZ <- crossprod(Z)
p <- ncol(Z)
W <- solve(ZtZ + lambda*diag(p))
bias <- c((W %*% ZtZ - diag(p)) %*% gamma)
varia <- sigmasq * diag( crossprod(Z %*% W))
list(bias = bias, variance = varia, mse = sum(bias^2 + varia))
}
set.seed(9876)
# Create fake data
Z <- matrix(rnorm(n = 20L*50L, mean = 0, sd = 1), ncol = 20L)
# Center and renormalize Z
Z <- apply(Z, 2, scale)
# Create coefficient vector
gamma <- c(rep(0, 10L), runif(10L))
# Create sequence of lambda and matrix to store results
nlam <- 401L
lambda_seq <- seq(0, 100, length = nlam)
mse <- matrix(0, nrow = nlam, ncol = 3)
gammahat <- matrix(0, nrow = nlam, ncol = ncol(Z))
for(i in 1:nlam){
# evaluate bias + variance for each lambda
mse_i <- mse_ridge(gamma = gamma, lambda = lambda_seq[i], Z = Z)
gammahat[i,] <- gamma + mse_i$bias
mse[i,1] <- sum(mse_i$bias^2)
mse[i,2] <- sum(mse_i$variance)
mse[i,3] <- mse_i$mse
}
# Plot the results as a function of lambda
matplot(lambda_seq, mse, type = "l", lty = 1,
bty = "l", xlab = expression(lambda), col = 3:1,
ylab = "Mean squared error decomposition")
abline(h = mse[1,3], lty = 2)
legend(x = "topleft", legend = c("sq. bias", "variance", "mse"),
col = 3:1, lty = 1, bty = "n")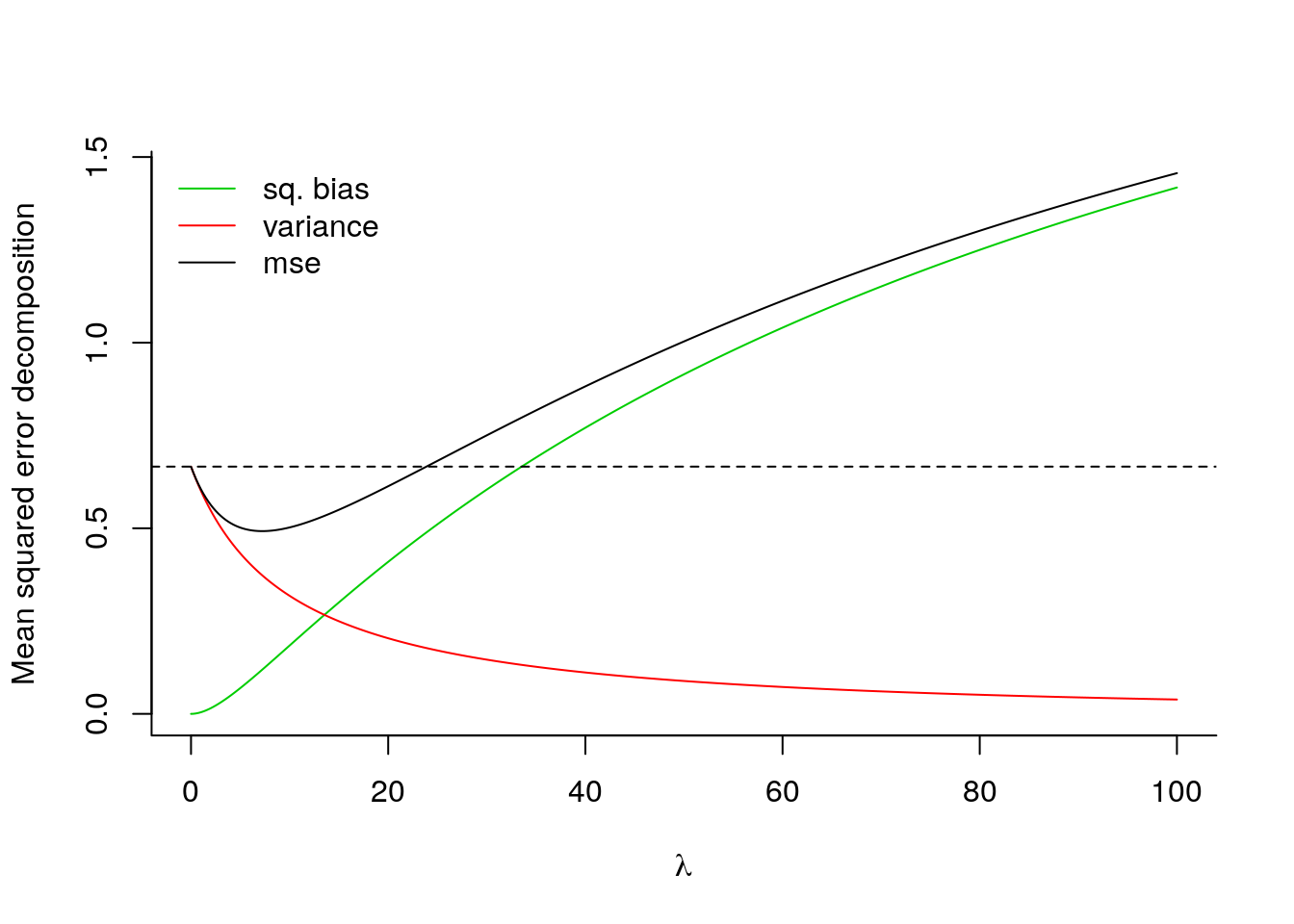
# Compute the value of lambda that minimizes MSE
lambdaopt <- optimize(f = function(x){
mse_ridge(gamma = gamma, lambda = x, Z = Z)$mse
}, interval = c(0, 30))$minimum
lambdaopt## [1] 7.227898We can also look at the path of coefficient values \(\hat{\boldsymbol{\gamma}}_{\mathrm{ridge}}^{\lambda}\) as a function of \(\lambda\):
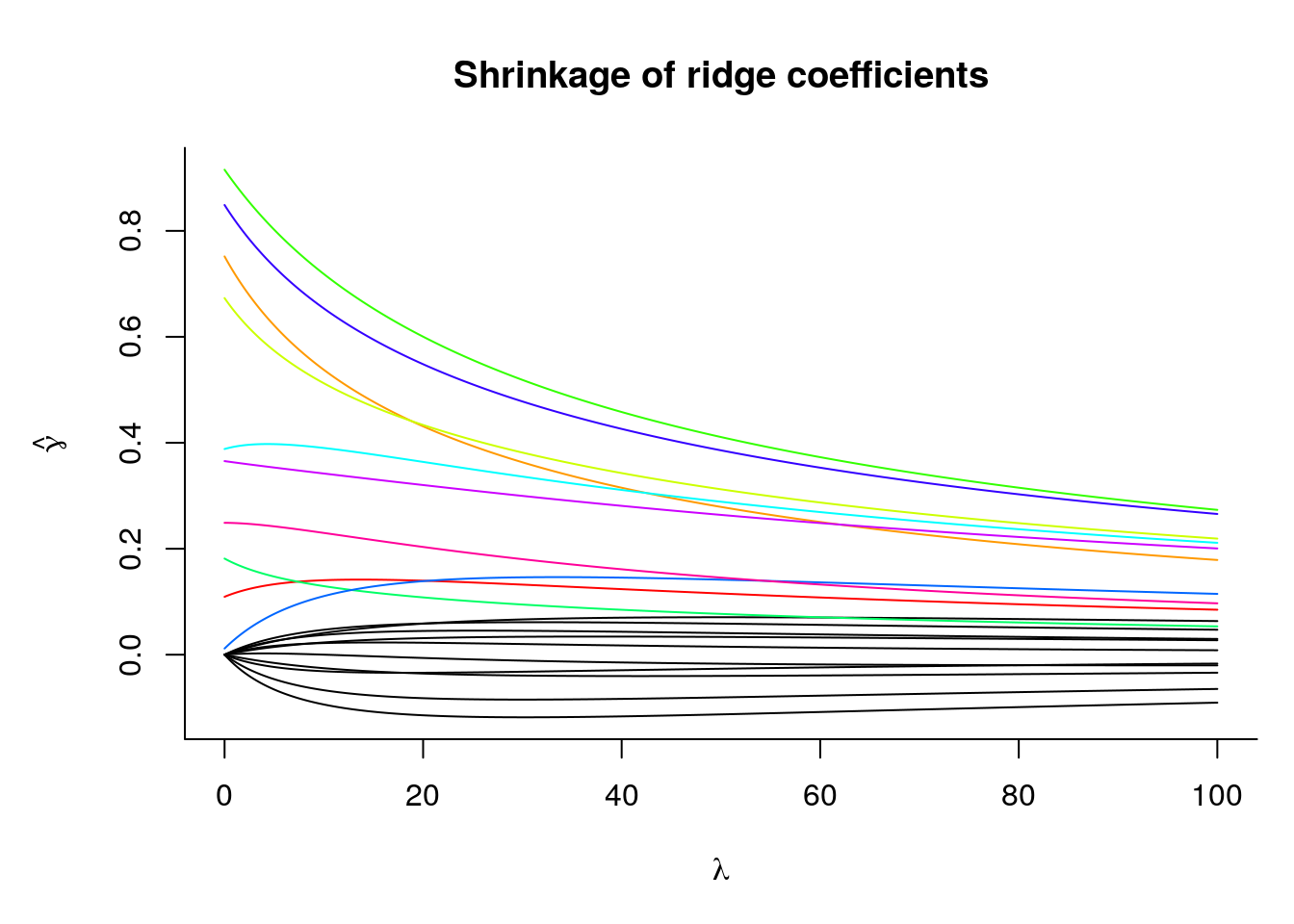
While the overall vector of coefficients are shrunk towards zero, the set of 10 first coefficients \(\boldsymbol{\gamma}\), which are exactly zero, stabilize around another value. Note that if we increase the penalty, from \(\lambda_1\) to \(\lambda_2\) with \(\lambda_1 < \lambda_2\), this does not necessarily imply that individual coefficient estimates decrease, i.e. \(|\hat{\beta}_j (\lambda_1)| \nleq |\hat{\beta}_j(\lambda_2)|\) even if \(\lambda_1 < \lambda_2\).
The coefficients \(\hat{\boldsymbol{\gamma}}^\lambda\) can be computed using an augmented linear regression, with response \((\boldsymbol{y}, \mathbf{0}_p)\) and regressor \([\mathbf{Z}^\top,\; \lambda^{1/2} \mathbf{I}_p]\). Alternatively, \[\hat{\boldsymbol{\gamma}}^\lambda = (\mathbf{Z}^\top\mathbf{Z} + \lambda \mathbf{I}_p)^{-1}\mathbf{Z}^\top\boldsymbol{y}.\]
We can also use the singular value decomposition of \(\mathbf{Z} = \mathbf{UDV}^\top\) to compute the coefficients\[\hat{\boldsymbol{\gamma}} = \sum_{j=1}^p \frac{d_j}{d_j^2+\lambda} \mathbf{u}_j^\top\boldsymbol{y} \mathbf{v}_j,\] where \(\mathbf{u}_j\) and \(\mathbf{v}_j\) are the \(j\)th column of \(\mathbf{U}\) and \(\mathbf{V}\), respectively. This is most useful for cross-validation where we want to change the value of \(\lambda\) repeatedly, as the SVD of \(\mathbf{Z}\) won’t change.
# Create response vector from model
y <- Z %*% gamma + rnorm(nrow(Z))
# Compute ridge coefficients
ridge_lm_coef <- function(y, Z, lambda){
Z <- scale(Z)
c(solve(crossprod(Z) + lambda*diag(ncol(Z))) %*% crossprod(Z, y - mean(y)))
}
lambda0 <- 2
#Compare coefficients obtained from fitting using augmented regression
augmy <- c(y - mean(y), rep(0, ncol(Z)))
augmZ <- rbind(Z, sqrt(lambda0)*diag(ncol(Z)))
augmlm <- lm(augmy ~ -1 + augmZ)
isTRUE(all.equal(as.vector(augmlm$coef),
ridge_lm_coef(y = y, Z = Z, lambda = lambda0)))## [1] TRUE